1. MolVision
VLMs for Molecular Property Prediction
[Project Page] [Code] [PDF]
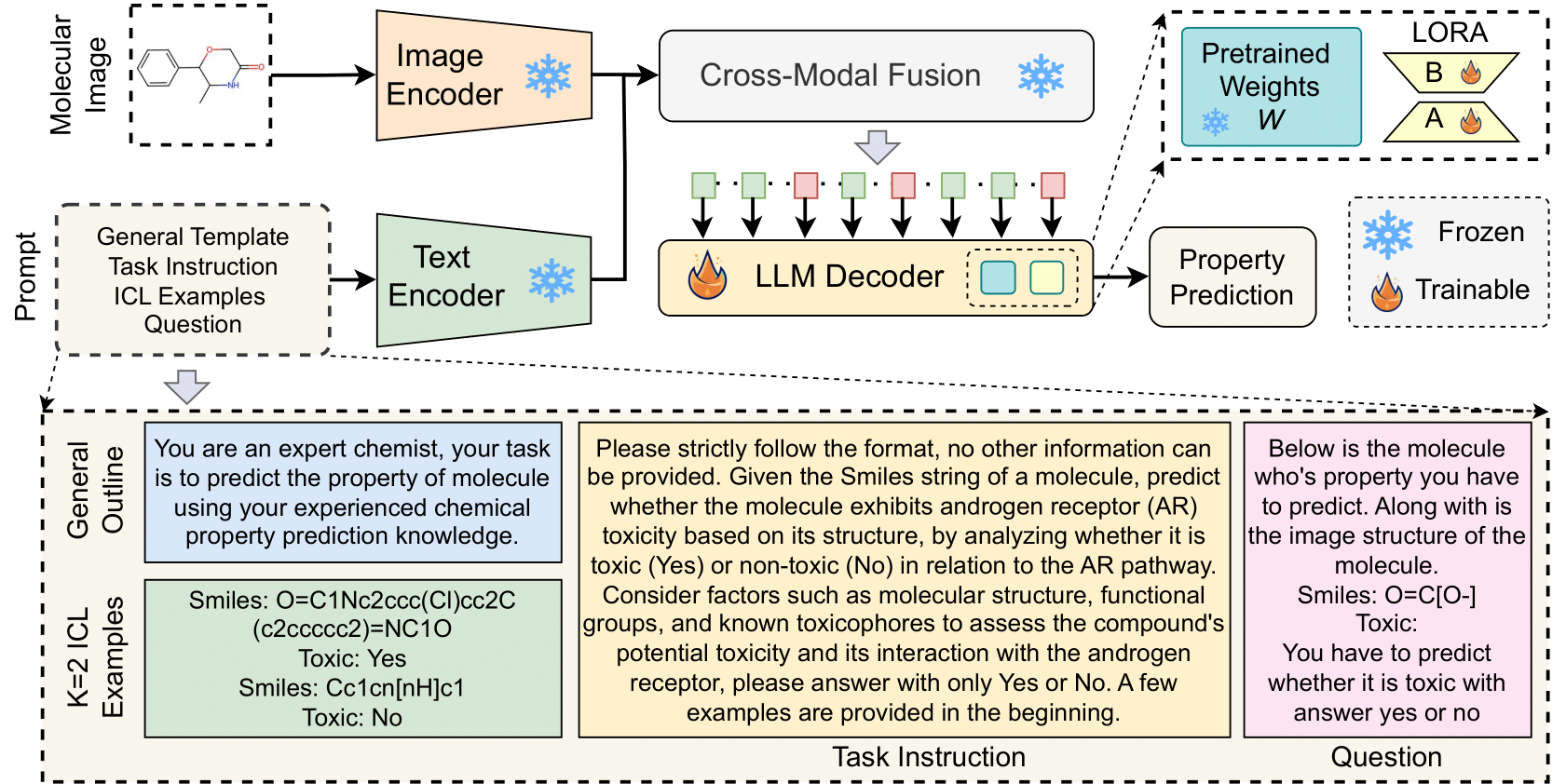
Molecular property prediction is a fundamental task in computational chemistry with critical applications in drug discovery and materials science. While recent works have explored Large Language Models (LLMs) for this task, they primarily rely on textual molecular representations such as SMILES/SELFIES, which can be ambiguous and structurally less informative. In this work, we introduce MolVision, a novel approach that leverages Vision-Language Models (VLMs) by integrating both molecular structure as images and textual descriptions to enhance property prediction. We construct a benchmark spanning ten diverse datasets, covering classification, regression and description tasks. Evaluating nine different VLMs in zero-shot, few-shot, and fine-tuned settings, we find that visual information improves prediction performance, particularly when combined with efficient fine-tuning strategies such as LoRA. Our results reveal that while visual information alone is insufficient, multimodal fusion significantly enhances generalization across molecular properties. Adaptation of vision encoder for molecular images in conjunction with LoRA further improves the performance (Adak et al., 2025).
🧬 MolVision - Characteristics and Statistics
Characteristics of MolVision
- Multimodal Integration: MolVision combines skeletal structure images with SMILES representations for molecular property prediction
- Diverse Datasets: It includes Ten datasets covering various molecular properties and complexities
- Evaluation Scenarios: Assessing Vision-Language Models (VLMs) under zero-shot, few-shot, Chain-of-Thought and fine-tuning conditions
- Comparative Analysis: Benchmarking Two closed source and Seven Opensourced different VLMs to analyze their effectiveness in computational chemistry
Statistics of MolVision
| Category | Details |
|---|---|
| Number of Datasets | 10 datasets: BACE-V, BBBP-V, HIV-V, Clintox-V, Tox21-V, Esol-V, LD50-V, QM9-V, PCQM4Mv2-V, Chebi-V |
| Dataset Composition | Includes skeletal structure images and corresponding SMILES strings |
| Model Evaluation | Two Closed Source and Seven OpenSourced Vision-Language Models evaluated |
| Performance Metrics | Measured across zero-shot, few-shot, Chain-of-thought and fine-tuning scenarios |
📋 Overview
This tool enables automated inference using OpenAI’s GPT-4 vision models on chemical datasets containing molecular images and associated questions/prompts. It’s particularly useful for:
- Molecular property prediction
- Chemical structure analysis
- Multi-modal AI evaluation on chemistry datasets
- Batch processing of vision-language tasks in chemistry